Tools/Suggestions
CMS Unix (lxplus) tools
Finding files using DAS CLI
The following python script takes a list of MC samples (dataset names) and lists all the files available on DAS using the command line interface. Ifdasgoclient is not available, run it from a CMSSW environment.
import os, sys
os.system('voms-proxy-init --voms cms')
print('Proxy generated. Investigating samples ..')
das_names = [
"/VLLSinglet_M-100_13TeV_TuneCUETP8M1-pythia8-madgraph/RunIISummer16NanoAODv7-PUMoriond17_Nano02Apr2020_102X_mcRun2_asymptotic_v8-v1/NANOAODSIM"
]
for name in das_names:
query = '"file dataset='+name+'"'
processline = "dasgoclient -query="+query
print('\nFiles for '+name.split('/')[1]+':')
#print(processline)
#break
os.system(processline)
Directly downloading files using xrdcp
Once I have a list of files I want to download, which can be made by using the previously tool, or manually picking up files from DAS-GUI, I ran a python For loop to bring all those files in a local area. First, I move to my T3 storage location (T3_CH_CERNBOX) and generate proxy to access the files.
[lxplus] cd /eos/user/p/phazarik
[lxplus] voms-proxy-init --rfc --voms cms #asks for PEM password
The text file containing the list of files looks like the following.
/store/mc/Run3Summer22NanoAODv12/TTto2L2Nu_TuneCP5_13p6TeV_powheg-pythia8/NANOAODSIM/130X_mcRun3_2022_realistic_v5-v2/2540000/03a1a21f-6bd8-4009-81c8-b09958600e8d.root
/store/mc/Run3Summer22NanoAODv12/DYto2L-4Jets_MLL-50_TuneCP5_13p6TeV_madgraphMLM-pythia8/NANOAODSIM/130X_mcRun3_2022_realistic_v5-v2/2520000/4069d292-5674-4b8f-9af1-1b45e34ffca8.root
/store/mc/Run3Summer22NanoAODv12/WZto3LNu_TuneCP5_13p6TeV_powheg-pythia8/NANOAODSIM/130X_mcRun3_2022_realistic_v5-v2/50000/19bc60ef-36f7-4fe7-bedd-55a966dcc250.root
/store/mc/Run3Summer22NanoAODv12/ZZto4L_TuneCP5_13p6TeV_powheg-pythia8/NANOAODSIM/130X_mcRun3_2022_realistic_v5-v2/2520000/8551c790-72b1-4de9-87d1-c3d24a2fcc17.root
/store/mc/Run3Summer22NanoAODv12/WWto2L2Nu_TuneCP5_13p6TeV_powheg-pythia8/NANOAODSIM/130X_mcRun3_2022_realistic_v5-v2/2530000/45e0cc8a-b2f7-4515-9d17-f48e6f888cde.root
/store/mc/Run3Summer22NanoAODv12/TTtoLNu2Q_TuneCP5_13p6TeV_powheg-pythia8/NANOAODSIM/130X_mcRun3_2022_realistic_v5-v2/2520000/660979e9-a672-4654-aee2-aa4686dd3ca0.root
Once the proxy is generated successfully, I run the following python script to download the files to their respective directories.
import os
# Reading the list of files from list.txt
with open('list.txt', 'r') as file_list:
file_paths = file_list.readlines()
for line in file_paths:
# Getting rid of leading/trailing spaces
line = line.strip()
# Extracting the folder name
split_path = line.split('/')
folder_name = os.path.join(split_path[3], split_path[4]) # Example: Run3Summer22NanoAODv12/DYto2L-4Jets_MLL-50_TuneCP5_13p6TeV_madgraphMLM-pythia8
# Creating the folder if it doesn't exist
if not os.path.exists(folder_name):
os.makedirs(folder_name)
# Constructing the xrdcp command
input_file = f"root://cmsxrootd.fnal.gov//{line}"
command = f"xrdcp {input_file} {folder_name}/"
# Executing the xrdcp command
print(command)
os.system(command)
#print(f"Copied {input_file} to {folder_name}/")
#break
print("All files copied.")
Luminosity calculation from json
brilcalc is available in lxplus at ~/.local/bin/brilcalc. It takes a json file as input, which contains run-number and
lumisections in the following format.
{
"321975": [[591, 593], [597, 599], [717, 734] , [736]],
....
}
While running brilcalc, it is recommended (by LUM POG) to use a physics approved normtag as follows.
brilcalc lumi --normtag /cvmfs/cms-bril.cern.ch/cms-lumi-pog/Normtags/normtag_PHYSICS.json -u /fb -i [json file name]
More information can be found in the LUM POG TWiki page.
Cross-section calculation
I made this tool for calculation of cross-section in MC samples using GenXSecAnalyzer. It takes a nanoAOD DAS string as input, finds a parent miniAOD file using DAS CLI and runs the GenXSecAnalyzer script. Example use:
python3 find_xsec_fromDAS.py --dataset /WtoLNu-2Jets_TuneCP5_13p6TeV_amcatnloFXFX-pythia8/Run3Summer22NanoAODv12-130X_mcRun3_2022_realistic_v5-v2/NANOAODSIM --dryrun
Submitting CRAB jobs and bringing samples
The CRAB framework manages distributed computing tasks by packaging user-defined analysis codes with configuration files that specify input datasets and output locations. Upon submission, jobs are distributed across the WLCG using a scheduler for dynamic resource allocation. It monitors job status through a database, allowing users to track progress and retrieve output files and logs, ensuring integration with CMS data management for efficient data analysis. Detailed instructions can be found here. I inherited the CRAB setup from Arnab, and organized it a bit. It is available in this GitHub repository. I use it to skim nanoAOD files in DAS and dump them in T2_IN_TIFR before bringing them to our cluster. Go to my setup and follow these steps.
- Login to lxplus8.
- Be in a CMSSW environment. In my case, its CMSSW_13_0_13, but the exact version is not crucial. This is just needed for using RDataFrame and submitting crab jobs.
- Generate voms-proxy for CMS to access the files from DAS.
-
Clone the following repository in your work area.
git clone https://github.com/phazarik/crabSkimSetup.git -
Run
crab_script.shlocally to see if it can process the input file mentioned inPSet.py. This is basically running a python script (nanoRDF.py) which is designed to skim the input file using RDataFrame. This may take a while to run in a local lxplus area. -
If previous step runs smoothly and produces an output file, submit the crab job by running
crab_config.py. This submits a crab-job for one dataset and manages howcrab_script.shshould run remotely. In my case, I run this in bulk, and feed some parameters externally. The following is an example of how to run the setup for one job.crab submit crab_config.py General.requestName=nanoRDF_Run2_2017_UL_Rare_THQ General.workArea=submitted Data.inputDataset=/THQ_ctcvcp_4f_Hincl_TuneCP5_13TeV_madgraph_pythia8/RunIISummer20UL17NanoAODv9-106X_mc2017_realistic_v9-v2/NANOAODSIM # Parameters fed from outside: # 1. General.requestName : Name of the crab-job that appears during monitoring. # 2. General.workArea : The crab-job logs are dumped in this folder, which is later used to monitor progress. # 3. Data.inputDataset : Full DAS string of the input sample # Rest of the parameters are same for all jobs, and are defined inside crab_config.py
Once the crab-jobs are done, the output files can be brought to any other location for analysis, as discussed in the next section.
TIFR-T2 tools
Generating proxy for CMS [hack using lxplus]
As of August 2024, the cms-proxy fails to generate at the T3 area. That's why I am manually creating the proxy file in lxplus, and brining it to ui.indiacms.res.in. Before brining the file to T3, the file has to be transferred to an accessible ares in lxplus.
[lxplus] voms-proxy-init --voms cms --valid 168:00
[lxplus] cp /tmp/x509up_u139657 . #Give the correct proxy filename
This file is brought into T3 area using scp as follows.
[ui3] scp phazarik@lxplus.cern.ch:~/x509up_u139657 . #Give correct username
[ui3] realpath x509up_u139657 # copy this
[ui3] export X509_USER_PROXY=/grid_mnt/t3home/phazarik/x509up_u139657 #Give the copied path here
Accessing and copying files to T3 [in bulk] using xrdcp
TIFR-T2 uses an xrood file system (similar to lxplus). Some important links are given below.
- T2/T3 User Guide (managed by TIFR)
- CMS TWiki: Using Xrootd Service
The T2 filesystem does not allow me to do ls easily and wildcards are not allowed. I also can't use python features like
os.listdir(). I use python scripts to run shell scripts such as xrdfs se01.indiacms.res.in ls from the T3 area and
list all the root files in a given directory. The same way I run xrdcp for all of them via For loop. First, I need to list the exact paths
containing the root files by running findPathsT2.py as follows.
[ui] python3 findPathsT2.py /store/user/alaha/nanoRDFjobs
This gives me the full-path to all the subfolders that contain root files. Then, for each path, I run getFilesT2.py individually as shown below.
[ui] python3 getFilesT2.py base_directory_in_T2 output_folder_in_T3
In case T2>T3 transfer fails
In case some files fail during the transfer as mentioned in the previous section, make a list of the paths to the individual files. The text file should look like the following.
/store/user/alaha/nanoRDFjobs/SingleMuon/NanoRDF__20240916_095804/240916_075807/0000/ntuple_skim_3.root
/store/user/alaha/nanoRDFjobs/Muon/NanoRDF__20240916_095809/240916_075812/0000/ntuple_skim_102.root
/store/user/alaha/nanoRDFjobs/Muon/NanoRDF__20240916_095809/240916_075812/0000/ntuple_skim_109.root
/store/user/alaha/nanoRDFjobs/Muon/NanoRDF__20240916_095809/240916_075812/0000/ntuple_skim_119.root
/store/user/alaha/nanoRDFjobs/Muon/NanoRDF__20240916_095815/240916_075817/0000/ntuple_skim_13.root
Then, run bring_individual_files.py as shown.
[ui3] python3 bring_individual_files.py outdir
Deleting files from T2
Deleting stuff from T2 is tricky. rm only works for individual files. rmdir only works for empty directories. I came
up with the following method. The python file takes a list of directories in this format: /store/user/username/directory.
- Listing all the folders, subfolders and files.
- Dividing the list into two: one for files, one for directories.
- Deleting the individual files first, by looping over the list of files.
- Deleting the directories, starting from the deepest ones.
For this, use cleanT2.py and mention the directories you want to remove in the list inside.
IISER T3 cluster tools
Finding out NanoAOD branches and their types
I made this tool to checkout and compare the branch types of different NanoAOD formats. For a given input file of a particular NanoAOD structure, it finds out the "Events" branch which is relevant to us. Then looping over each branch, it accesses the name and leaves associated with the branch and prints out the type. The code can be found here.
Checking number of events (generated) in NanoAOD files
For luminosity calculation, the total number of generated events is required. In our cluster, we have a mixture of the original nanoAOD file and
skimmed nanoAOD files. In case of the original nanoAOD files, nEvtGen is simply the number of entries in the Event tree. For the skimmed files,
this number is kept in a branch called 'nevtgen'. (This branch is filled for each event, so this number is repeatedly filled for all the
entries; we just need to access the value at the first entry.) This number is needed in case we miss any file while transferring from DAS to our
cluster. Run calculate_True_nEvtGen.py that works for both
skimmed and original nanoAOD files.
Note: This uses pyROOT. So make sure that you are in a CMSSW environment, and a compatible python version (that supports
import ROOT) is used. The following is an example usage.
python calculate_True_nEvtGen.py /home/work/alaha1/public/RunII_ULSamples/2017/Rare
# Note that
# 1. It's using python2.
# 2. Full-path to the sample-directory is given, where it finds all the sub-directories.
Generating a MakeSelector based template to analyze NanoAOD files
For a given NanoAOD file, a MakeSelector() based template can be generated in th following way. Make sure to note down the NanoAOD
version, because this template may have some branch-mismatch issue with other versions. Read the ROOT file and do the following in the ROOT
prompt.
[root] TFile *f = new TFile("filename.root")
[root] gROOT->FindObject("Events")
[root] Events->MakeSelector("anaName") #Pick an analysis name
This should produce a template analyzer class named
anaName. The class is kept in a header file, and its functions including the event loop is run in a C file. An example of such a
template code with some additional features is available in this git repository:
phazarik:nanoAOD_analyzer. I modified the template to be compatible
with multiple NanoAOD versions.
Miscellaneous tools
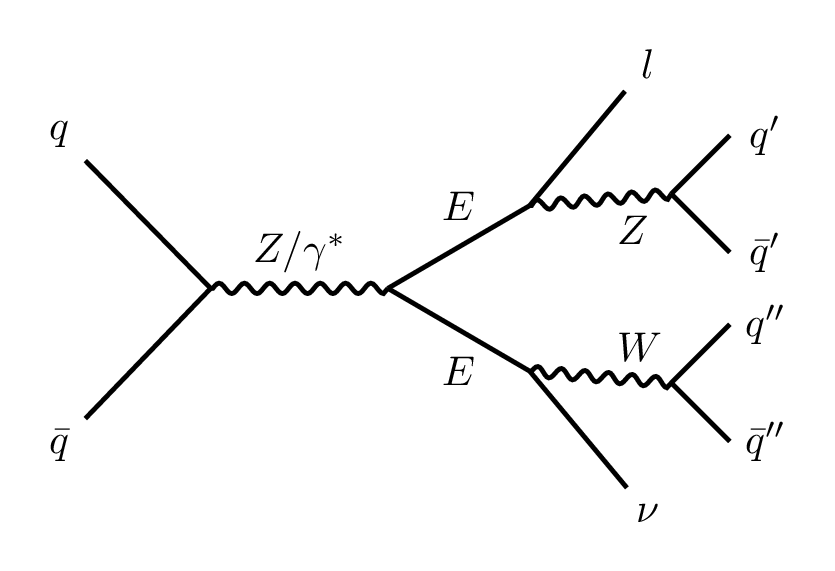
Drawing Feynman diagrams in LaTeX
When it comes to drawing Feynman diagrams in LaTeX using feynmf and TikZ-Feynman, the later is generally easier and more flexible to use. The
first one is older and requires a more complicated setup, while TikZ-Feynman works smoothly with modern LaTeX, producing high-quality diagrams
with more customization options. If you're just starting out, TikZ-Feynman is often the better choice because it’s simpler and integrates well
with other LaTeX features. A template for drawing Feynman diagrams using tikz-feynman package can be found
here. It already has some examples. It can be compiled using
pdflatex as follows.
pdflatex feynman-template.tex
Just make sure that you have the necessary LaTeX packages installed beforehand. I prefer to produce individual pdf files for each Feynman diagram and convert them into high quality png files. The following is an example of a VLL production diagram.
\documentclass[tikz,border=3mm]{standalone}
\usepackage{tikz-feynman}
\tikzset{every picture/.style={line width=1.1pt}}
\begin{document}
\begin{tikzpicture}[baseline={(current bounding box.center)}]
\begin{feynman}
\vertex (v1);
\vertex [right =1.5cm of v1] (v2);
%incoming vertices
\vertex [above left =1.5cm of v1] (i1) {\(q\)};
\vertex [below left =1.5cm of v1] (i2) {\(\bar{q}\)};
%internal vertices connected to the v2
\vertex at($(v2)+(1.2, +0.7)$) (b1);
\vertex at($(v2)+(1.2, -0.7)$)(b2);
%internal vertices connected to b1 and b2
\vertex at($(b1)+(1.2, +0.1)$) (c1);
\vertex at($(b2)+(1.2, -0.1)$) (c2);
%outgoing vertices
\vertex at($(b1) + (1.0, +1.2)$) (o1) {\( l \)};
\vertex [above right =0.7cm of c1] (o2);
\vertex [below right =0.7cm of c1] (o3);
\vertex [above right =0.7cm of c2] (o4);
\vertex [below right =0.7cm of c2] (o5);
\vertex at($(b2) + (1.0, -1.2)$) (o6) {\( \nu \)};
\diagram*{
%incoming lines
(i1) -- (v1) -- (i2);
%internal lines
(v1) --[boson, color=black, edge label = \({\color{black} Z/\gamma^*}\)] (v2);
(b1) -- (v2) -- (b2);
(b1) --[boson, edge label' = \(Z\)] (c1);
(b2) --[boson, edge label = \(W\)] (c2);
%outgoing lines
(o1) -- (b1);
(o2) -- (c1) -- (o3);
(o4) -- (c2) -- (o5);
(b2) -- (o6);
};
%labels (manually putting them here because too crowded)
\vertex at($(o2) + (+0.3, +0.0)$) (l2) {\(q^{\prime}\)};
\vertex at($(o3) + (+0.3, +0.0)$) (l3) {\(\bar{q}^\prime\)};
\vertex at($(o4) + (+0.3, +0.0)$) (l4) {\( q^{\prime\prime}\)};
\vertex at($(o5) + (+0.3, -0.0)$) (l5) {\( \bar{q}^{\prime\prime}\)};
\vertex at($(b1) + (-0.6, +0.0)$) (tau1) {\( E \)};
\vertex at($(b2) + (-0.6, -0.0)$) (tau2) {\( E \)};
\end{feynman}
\end{tikzpicture}
\end{document}
This example only requires the tikz-feynman package. The pdf output can be converted to png as follows.
# Note: Don't use the -alpha remove option if you want transparent png files.
convert -density 300 vll_production.pdf \
-colorspace RGB -quality 90\
-alpha remove -background white\
vll_production.png